Finding genes

Finding genes using gene-finding programs
One of the key goals of genomics research is to identify the genes responsible for a particular phenotype. There are several methods of doing this.
If genomic sequence is available, there are a number of "gene-finding" software programs which can identify candidate genes, mainly by identifying open reading frames (locating start and stop codons). However, due to differences among genomes, these programs are not 100% accurate and need to be adjusted for specific species.
Some of the programs available are Genescan (from MIT), ORF Finder (from NCBI) and TigrScan (from TIGR).
Finding genes using other known genes
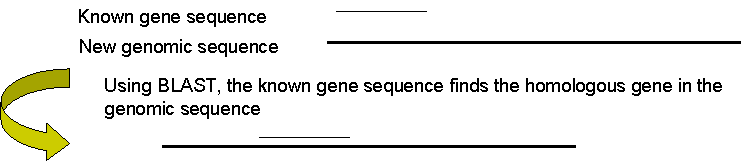
If genome sequence is not available, another method of identifying genes is to exploit sequences from other species which have already been identified as genes, and use these to search for corresponding genes in the new sequence (this is possible due to the high level of conservation of genes among most organisms).
For example, the known gene sequence could be compared to the new genomic sequence using BLAST.
Other methods of finding genes
Generating ESTs, which produces large numbers of short genic sequences.
Generating full-length cDNA clones. As these are longer than EST sequences, the necessary libraries are more difficult to construct and other sequencing strategies are required.
Identification of genes via mutagenesis. This approach is sometimes termed "reverse genetics". Possible strategies are discussed below.
Identification of genes via mutagenesis
"Gene knock-outs" in which the insertion of aT-DNA element or a transposon inactivates the gene. The sequence of the T-DNA/transposon is used to detect which gene has been disrupted.
"Targeted induced local lesions in genomes (TILLING)", in which chemical mutagenesis induces point mutations, and these are identified by screening PCR products.
"RNA interference (RNAi)", where all members of a particular gene family are "silenced" (turned off) by the transgenic insertion of a specific double-stranded RNA.
The Nobel Prize in Physiology or Medicine for 2006 was awarded jointly to Andrew Z. Fire and Craig C. Mello for their discovery of "RNA interference -- gene silencing by double-stranded RNA."





