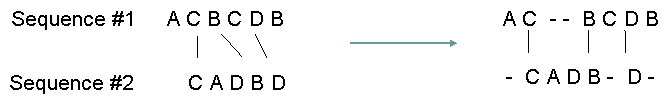Sequence alignment

In this section we will consider:
Once you have a sequence (or set of sequences), the next step is usually to establish whether there are any similar sequences already in the database, whether they have been assigned a function, and what else, if anything, is known about them. This first step aligns your sequence against all the other sequences in the database.
Below is a simplified example of alignment (taken from Schneider & La Rota, 2000). Note that gaps (signified by "-") are introduced by the algorithm to allow for insertions or deletions that have occurred in the sequences.