

Breaking linkage drag, contd.
A major advantage of using advanced backcross populations for QTL mapping is that each time a cross is made back to the recurrent parent (usually the cultivated variety, if the other parent is a wild relative), less of the donor genome remains.
Once a locus in the segregating population becomes “fixed” (homozygous) for the recurrent parent allele (AA), you no longer need to check it with markers, because it can only stay homozygous in each subsequent backcross generation.
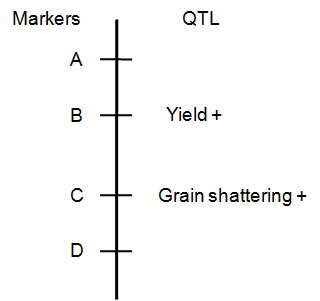
Therefore if your recurrent parent is AA, and in your segregating population Marker A and Marker D are AA, after a cross back to the recurrent parent, you only need to check the progeny with Markers B and C, and hopefully find a plant that is Aa only at B (linked to the yield QTL is) and now AA at Marker C (has lost the bad QTL due to recombination).

