Summary
This tutorial describes how to design a field trial to evaluate 32 (entries) germplasm for 7 phenotypic traits replicated three times at 4 locations. The experimental design is a resolvable incomplete block design, specifically an alpha lattice design.
Restore Example Data
Screenshots and activities in this tutorial build upon preloaded example germplasm.
- If you are not following the maize tutorials in sequence, restore the Maize Tutorial database (.sql) with example data.
Restoration File: Maize Database with 3 Germplasm Lists (.sql)
Design Trial
Trial Details & Settings
- From Manage Trials, select Start a New Trial.

- Establish basic trial details. Leave the default settings and establish a name and description.
- Name: Performance Trial
- Description: 2017
- Select Add to add management details. Select PI_NAME from dropdown menu.
- PI_NAME: Self
- Save.
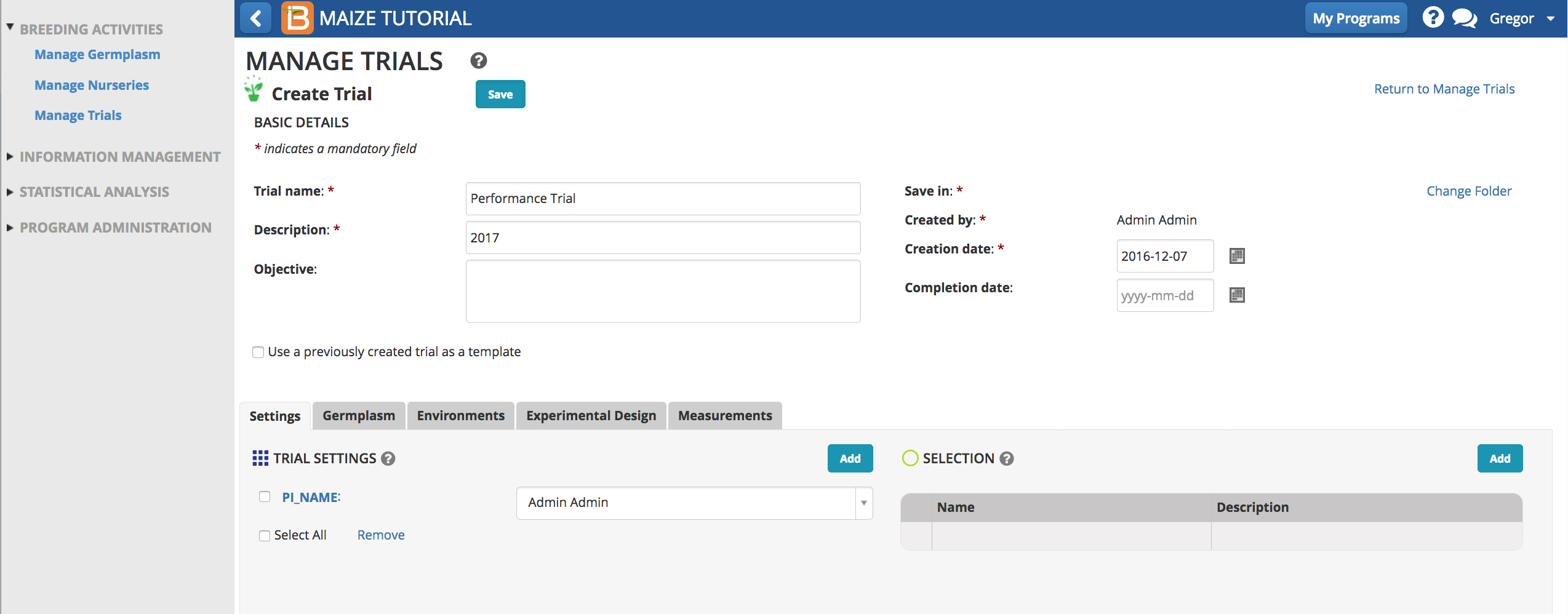
- Save trial in 2017 folder.
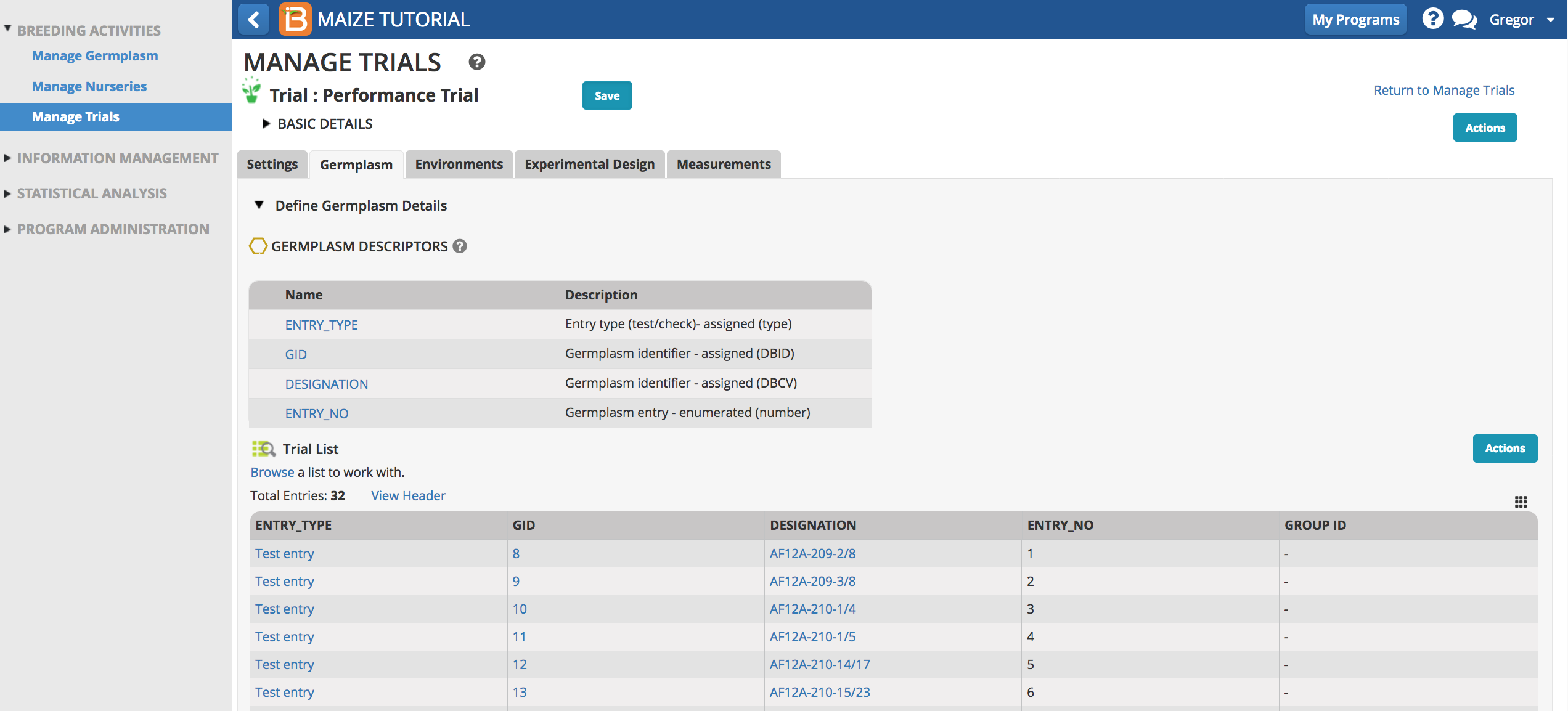
Germplasm
- Browse for a germplasm list.
- Select the Trial Germplasm list.
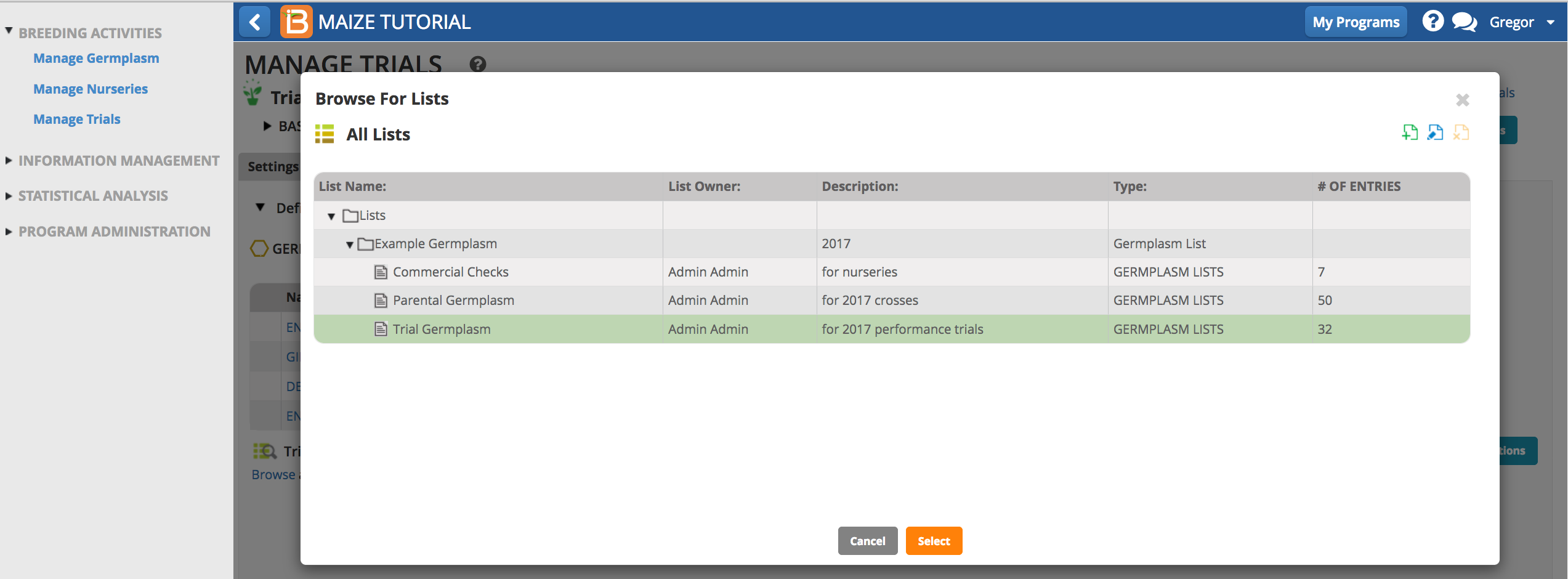
The 32 germplasm entries are now loaded into the trial.
Environments
- Specify four environments for the trial and select OK. Four environments will now populate the screen below.
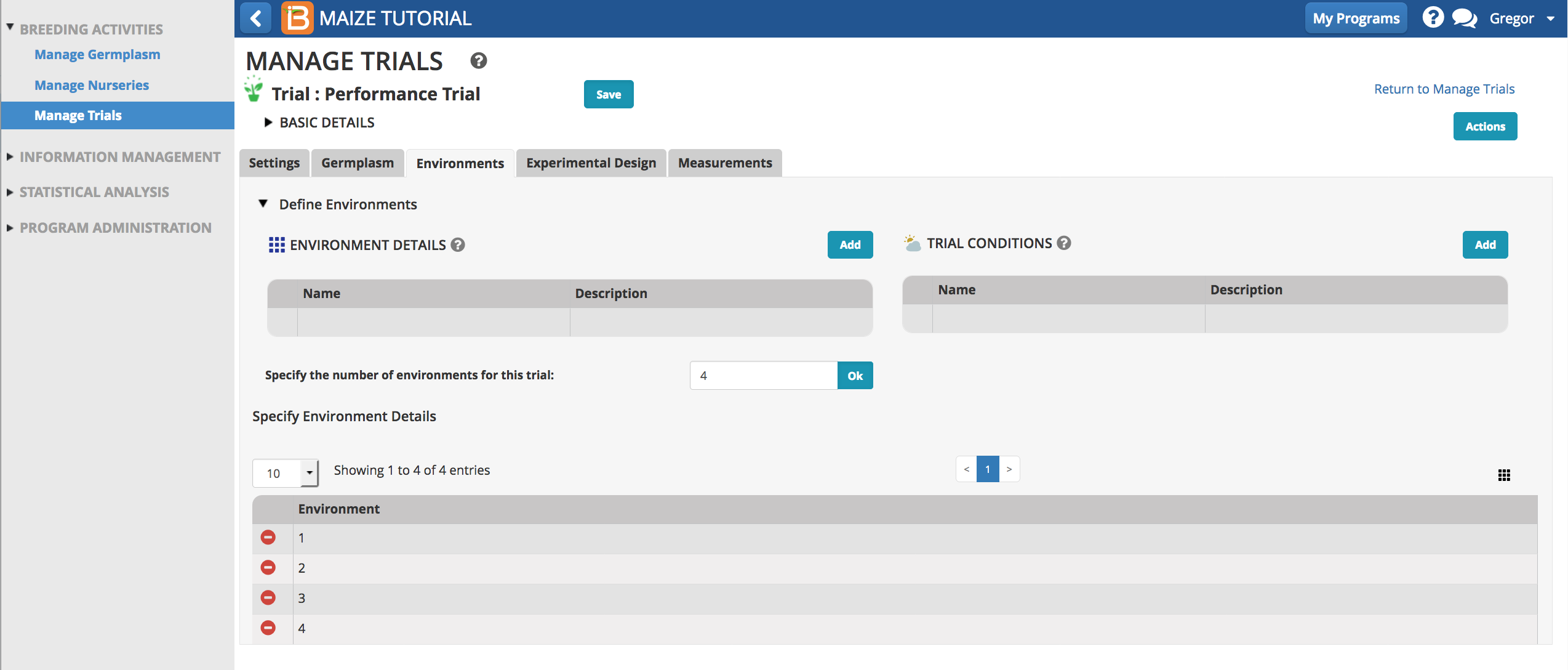
The 4 environments correspond to the following location names:
- ?Environment 1: Agua Fria - (AF)
- Environment 2: Sabana Del Medio - (SDM)
- Environment 3: Jutiapa - (JUT)
- Environment 4: Tlaltizapan - (TLA)
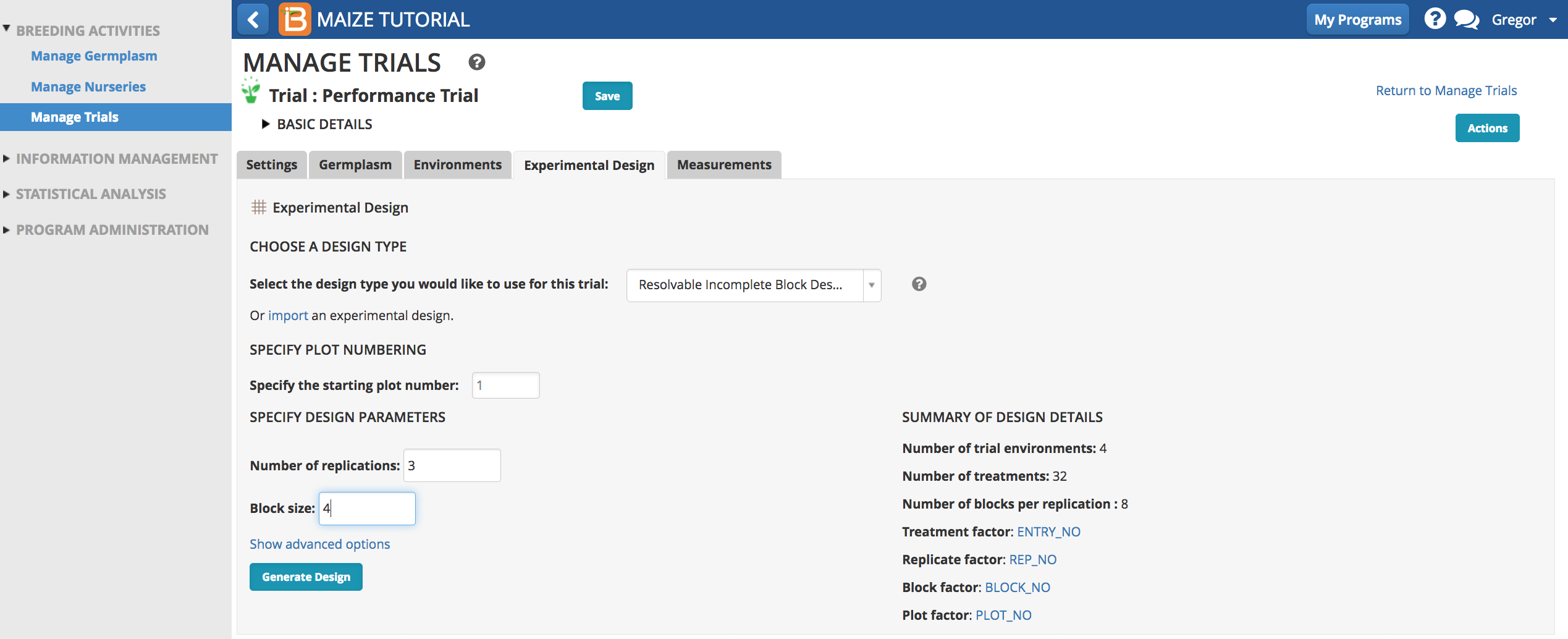
Experimental Design
The BMS supports the randomization of several types of experimental design. This experiment has a resolvable incomplete block design, where all germplasm are replicated 3 times at 4 locations containing 4 incomplete blocks. The advantage of this design over a randomized complete block is the ability to control for environmental heterogeneity within a location. For this design the number of germplasm entries must be evenly divisible by the size of the block (32 germplasm / 4 plots per block = 8 blocks per replicate).
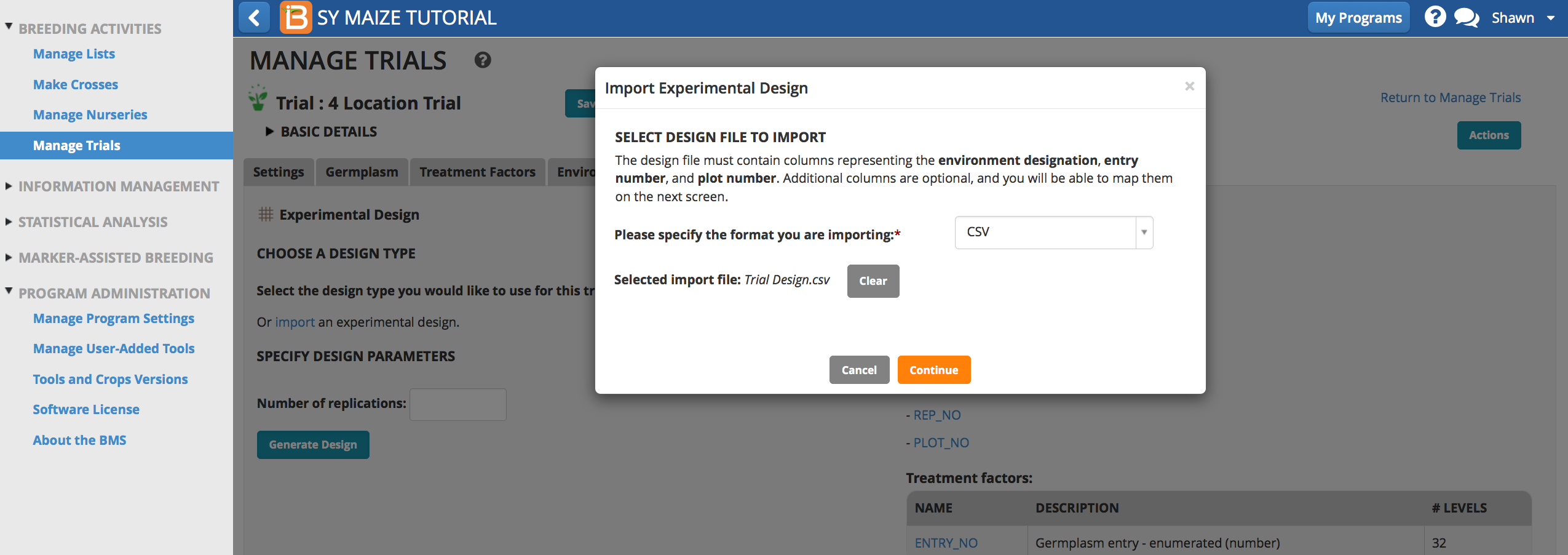
Import Trial Design
- Every design generated will have a different randomization. For the purpose of following the tutorials, upload an experimental design file to match trial design to preformatted data import files. Select import experimental design from Actions Menu and browse to the import file. Select Continue.
Experimental Design File: Trial Design (.csv)
- Disregard the warning message. This experimental design was generated by the BMS, and will be compatible with BMS analysis tools. There are no unmapped variables. Select Next.
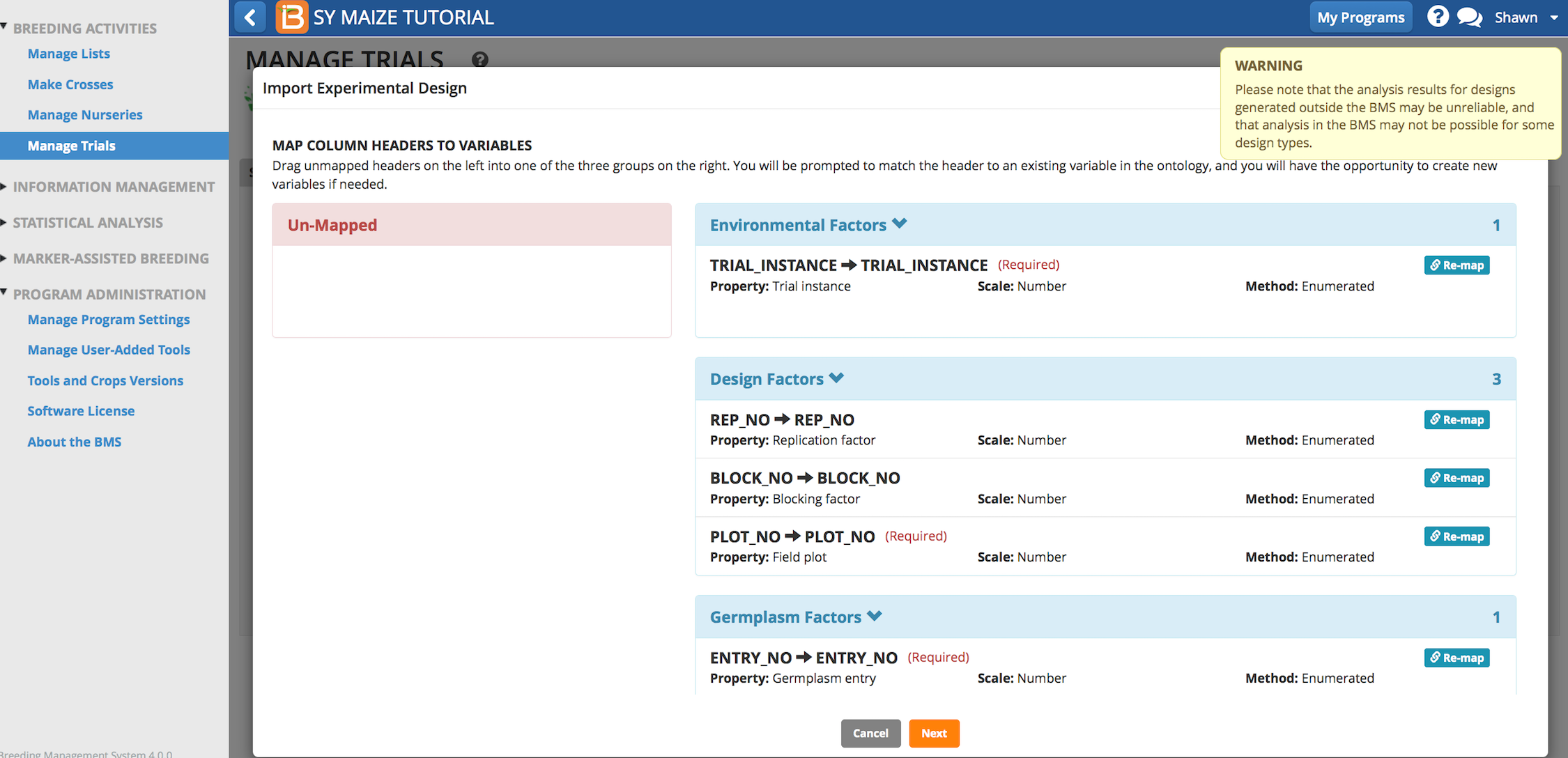
- If you have generated a trial design, select Yes to overwrite the previous design.
- Review design details and select Finish at the bottom of the pop-up window.
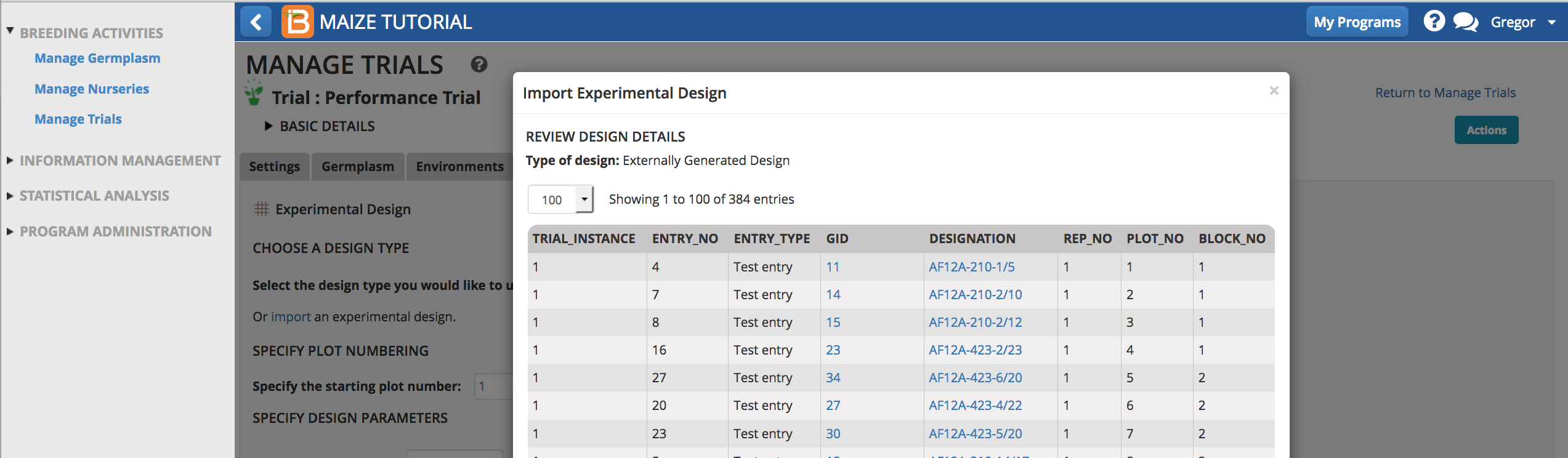
- Review the import details and select the measurements tab.
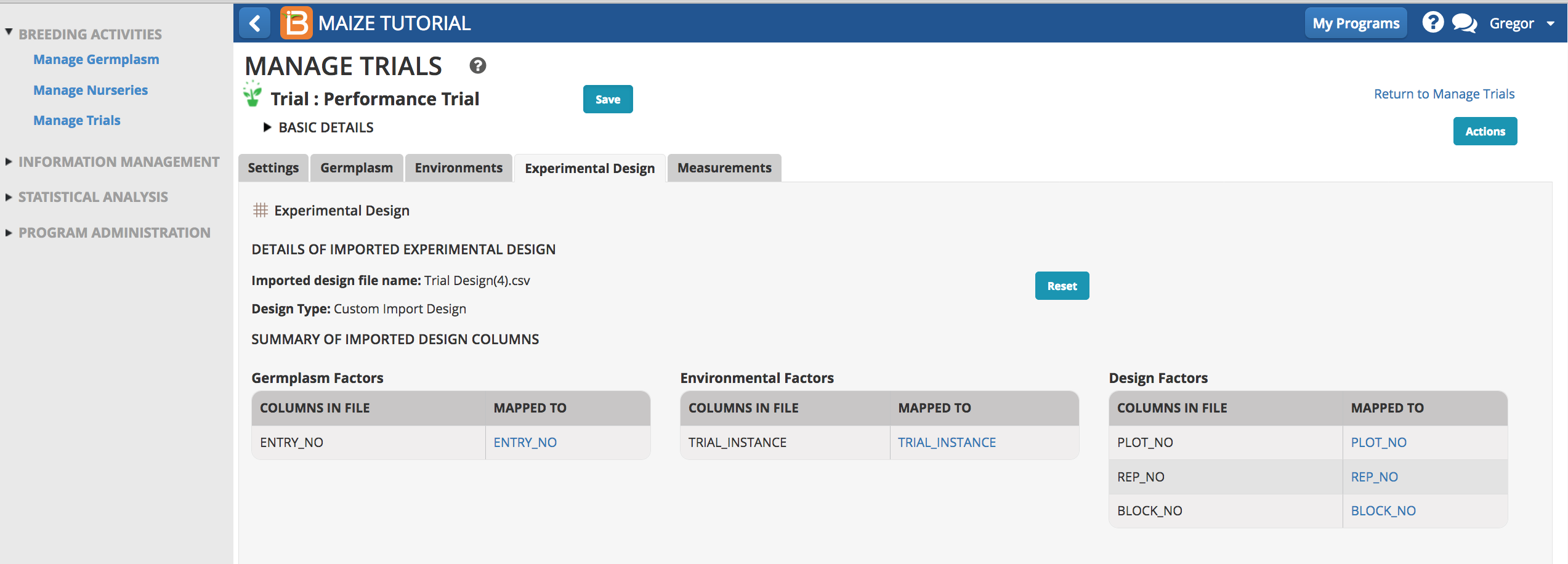
Measurements
Once the trial design is imported, the measurements table is populated with independent variables and the dependent trait variables can be specified.
- Add phenotypic traits by selecting Add.
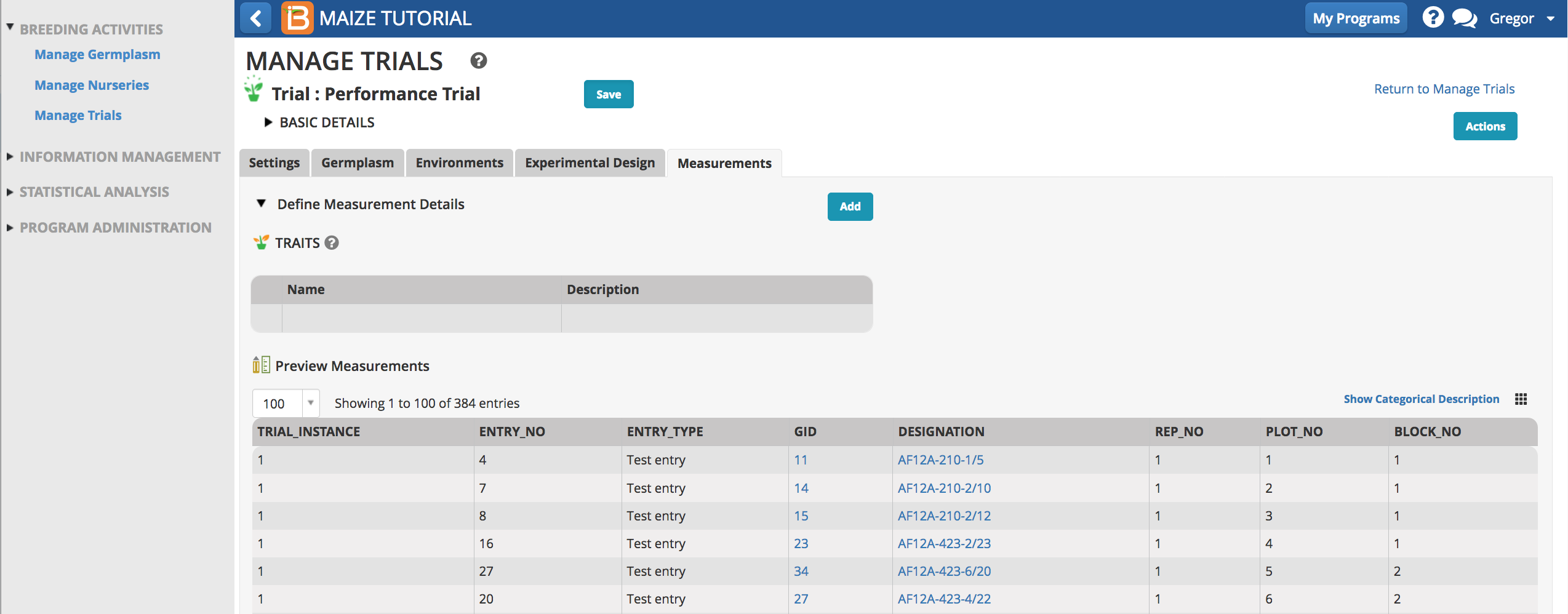
- Search for the six traits by typing details of the individual traits in the selection bar. For example typing the first few letters of ‘Yield’ reveals all of the related measurements.
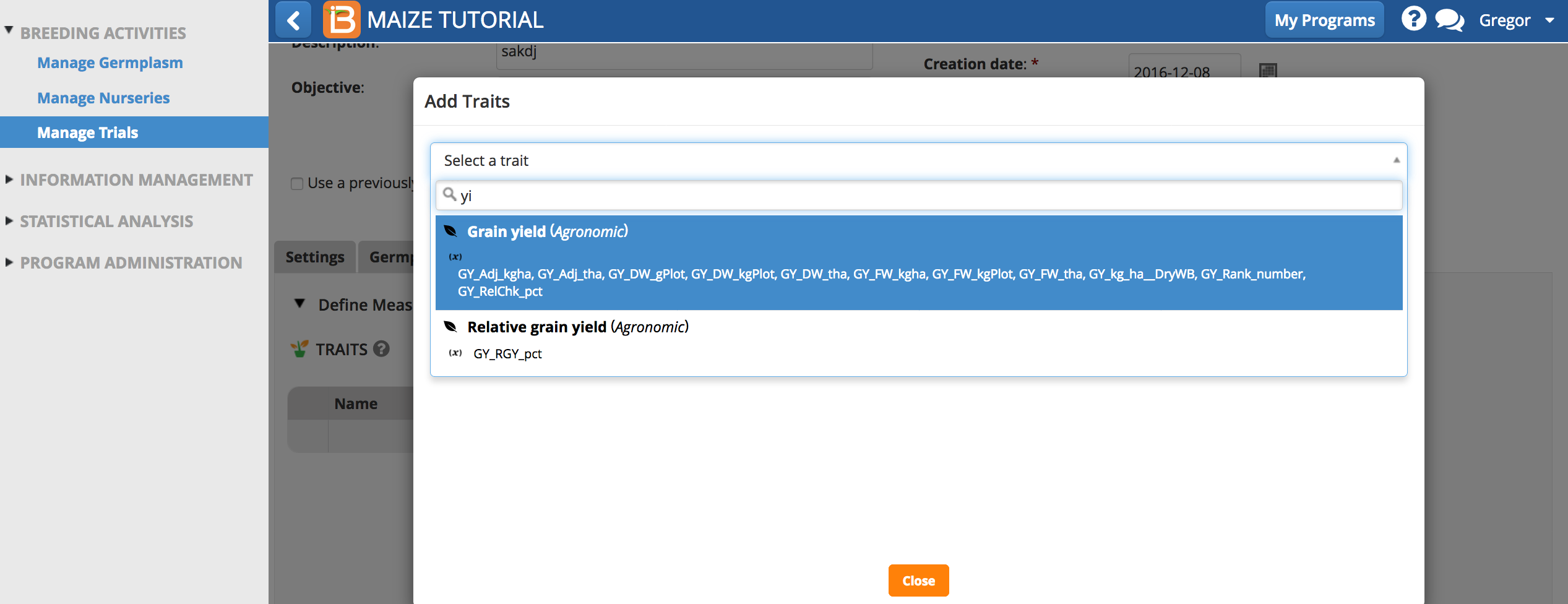
- Add the trait, GY_FW_kgPlot, grain yield by fresh weight in Kg/plot to the trial. Repeat by adding other traits of interest.
- Grain Yield Fresh Weight (GY_FW_kgPlot,): Grain yield BY FW GY - Measurement IN Kg/plot
- Grain Moisture (GMoi_NIRS)_pct: Grain smoiture BY NIRS Moi - Measurement IN %
- Plant Height (PHTsIB_M_cm): Plant height to the insertion of the first tassel branch BY PHTsIB - Measurement IN Cm
- Ear Height (EH_M_cm): Ear height BY EH - Measurment IN Cm
- Grain Yield Dry Weight (GY_DW_gPlot,): Grain yield BY DW GY - Measurement IN G/plot
- Anthesis Date (DTA_days_obs): number of days from sowing to when 50% of the plants shed pollen.
- Save the trial.

Plant Trial
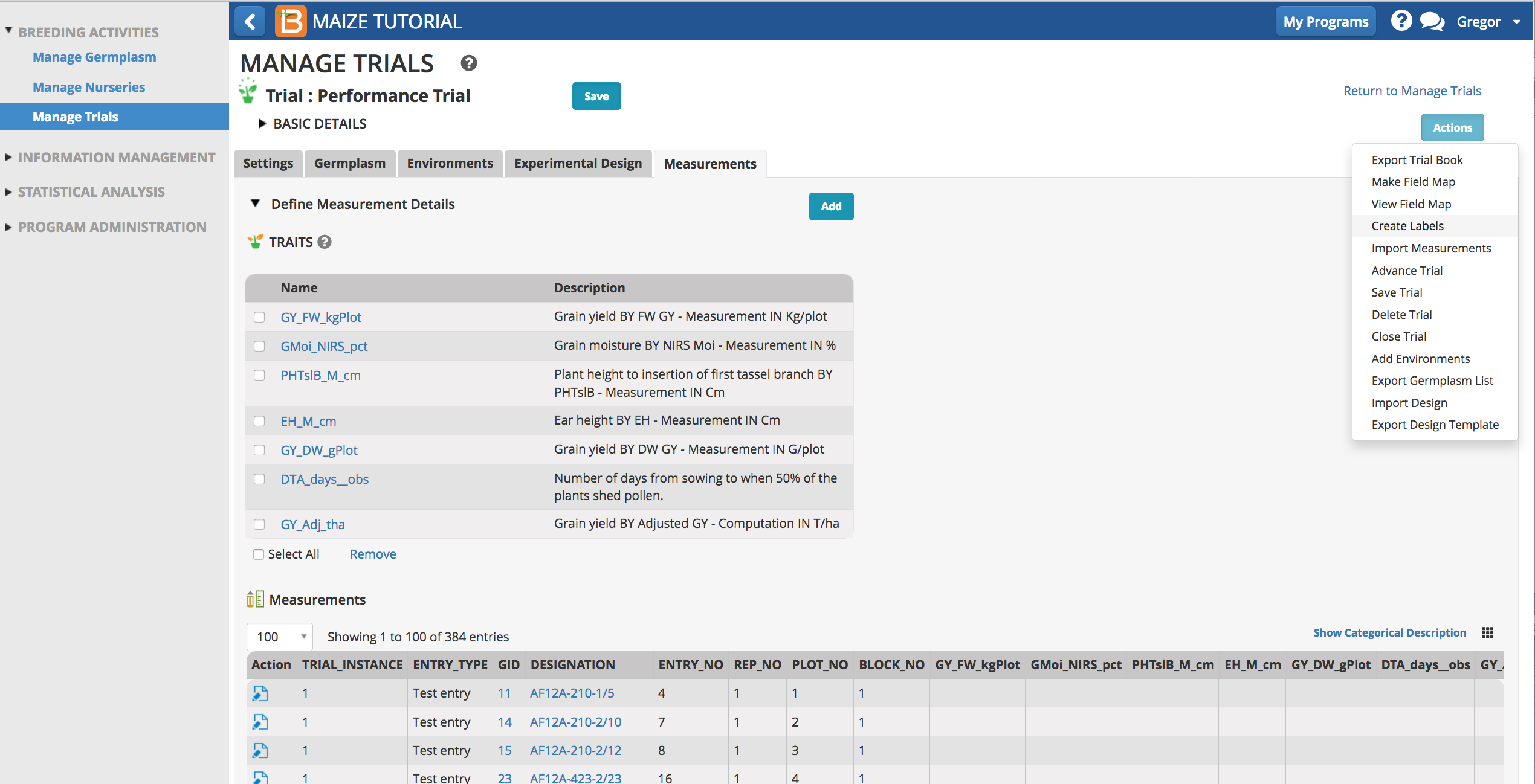
Trial design is complete when the measurements table is saved. Trial labels are available for seed packing and plot marking.
- Select Create Labels.
- Choose Formatted PDF Label Sheets.
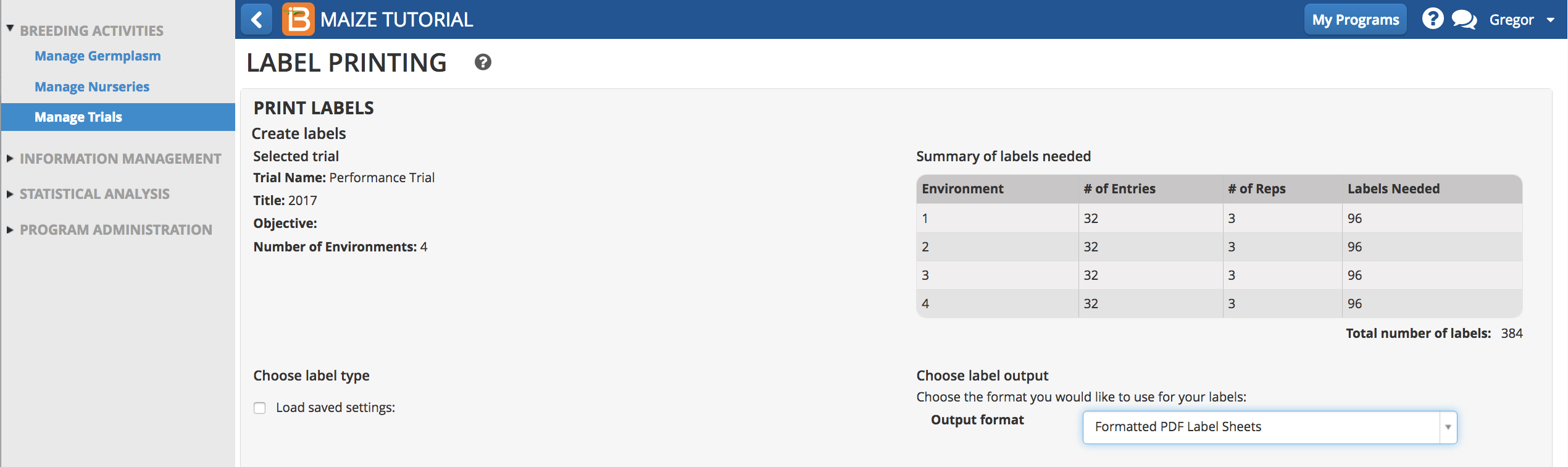
- Choose details to include on label text.
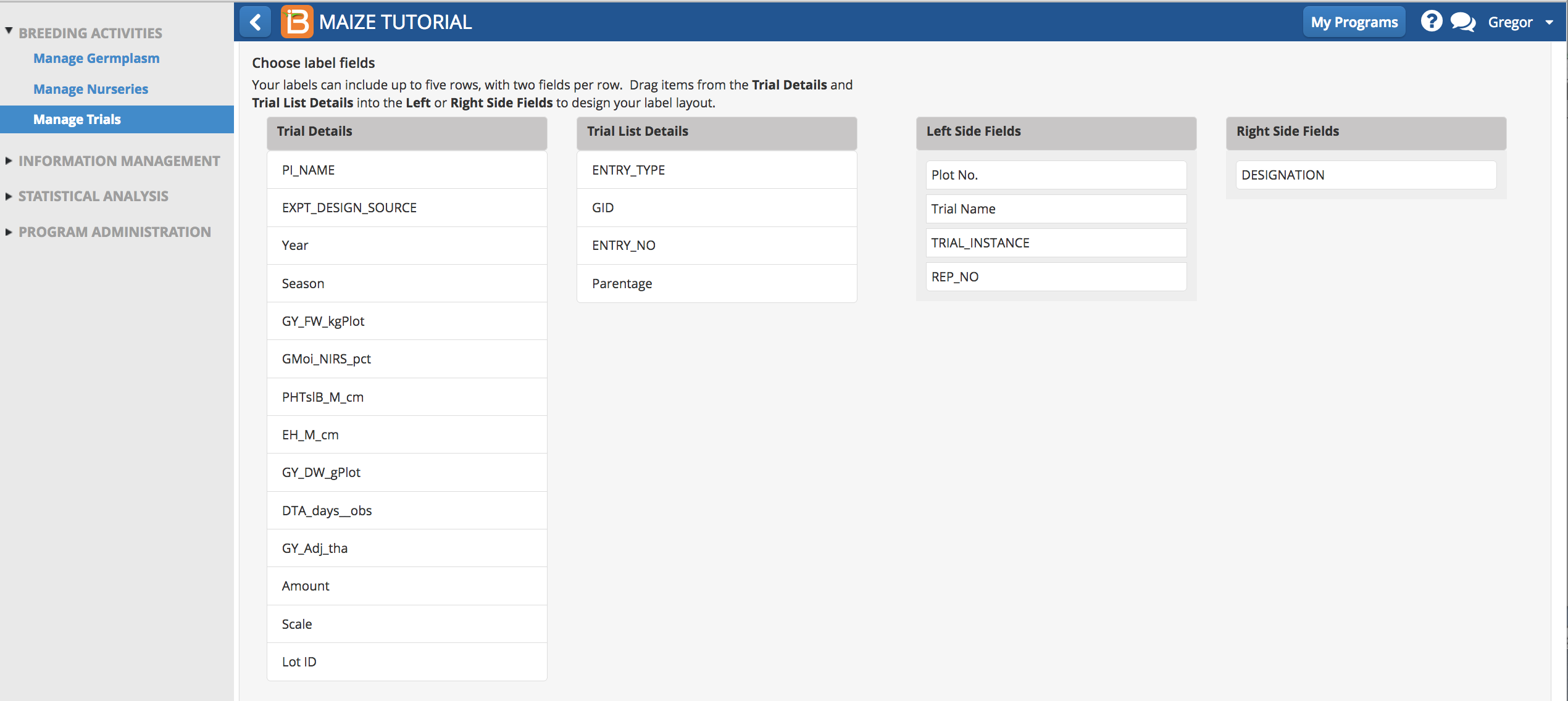
- Choose details to include in bar code. Export Label.
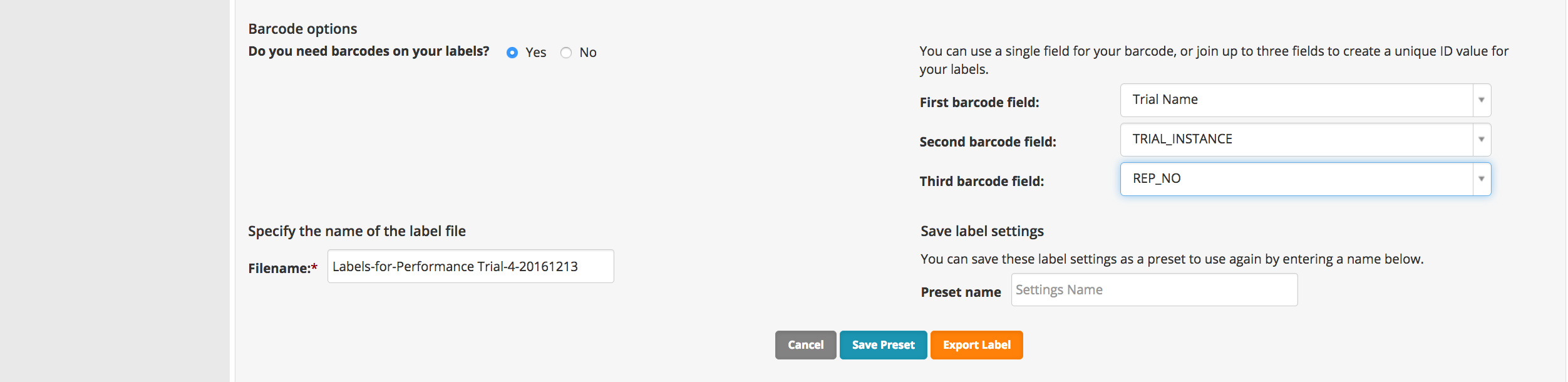
- Review the labels (.pdf). The set of trial labels can be printed twice to label (1) the seed destined for the trial and (2) the trial plots in the field.

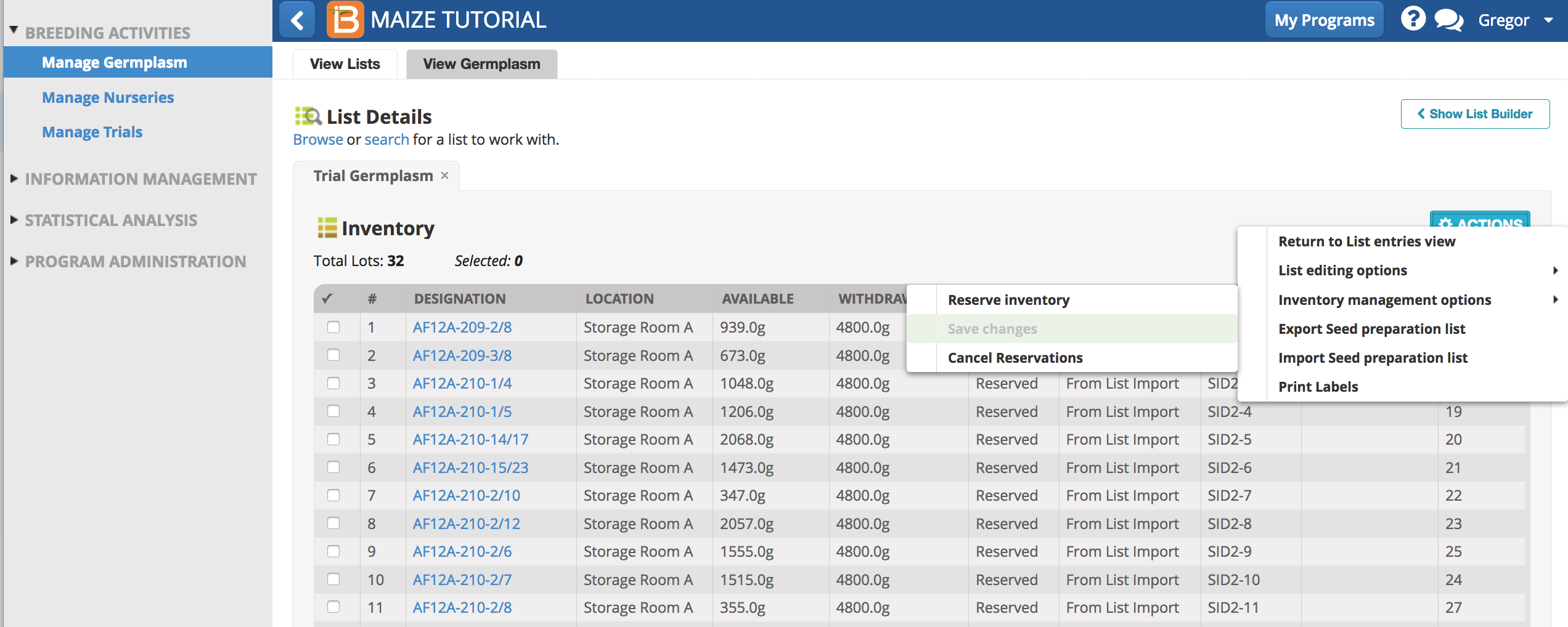
Reserve & Withdraw Trial Germplasm
If 400g of seed are needed per trial plot, and each trial germplasm is replicated 3 time over 4 environments - each trial germplasm entry requires a reservation and withdrawal of 4800g of seed. Trial labels can be used to track and match packaged seed to appropriate plots.
- From Manage germplasm browse and select the Trial Germplasm list. Open the list in Inventory View.
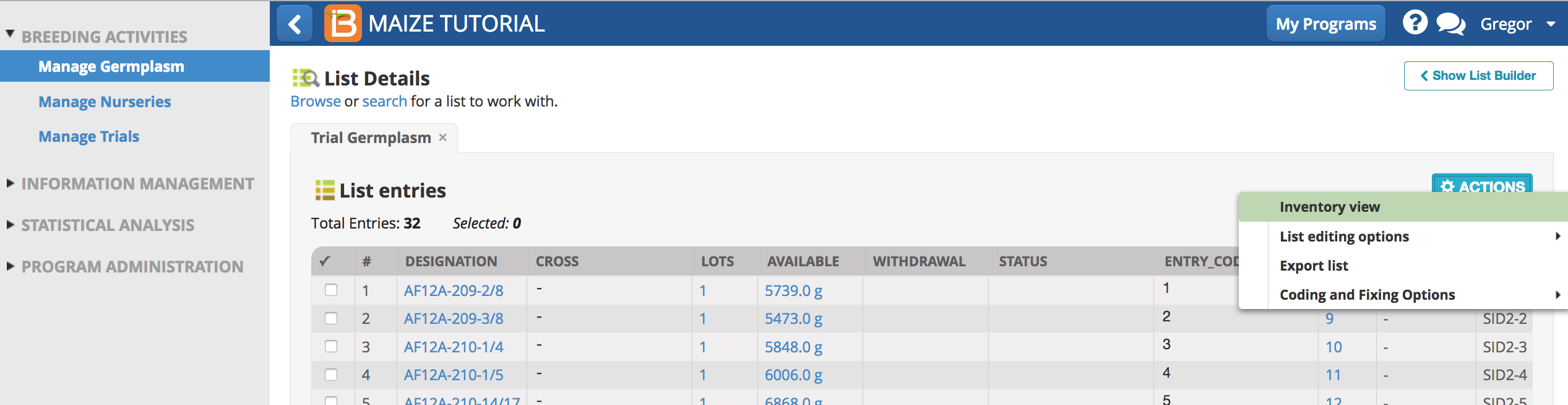
- Select al 32 entries and Reserve Inventory.

- Enter 4800g for seed amount. Select Commit seed withdrawal on saving to complete the reservation/withdrawal in one step. Finish.
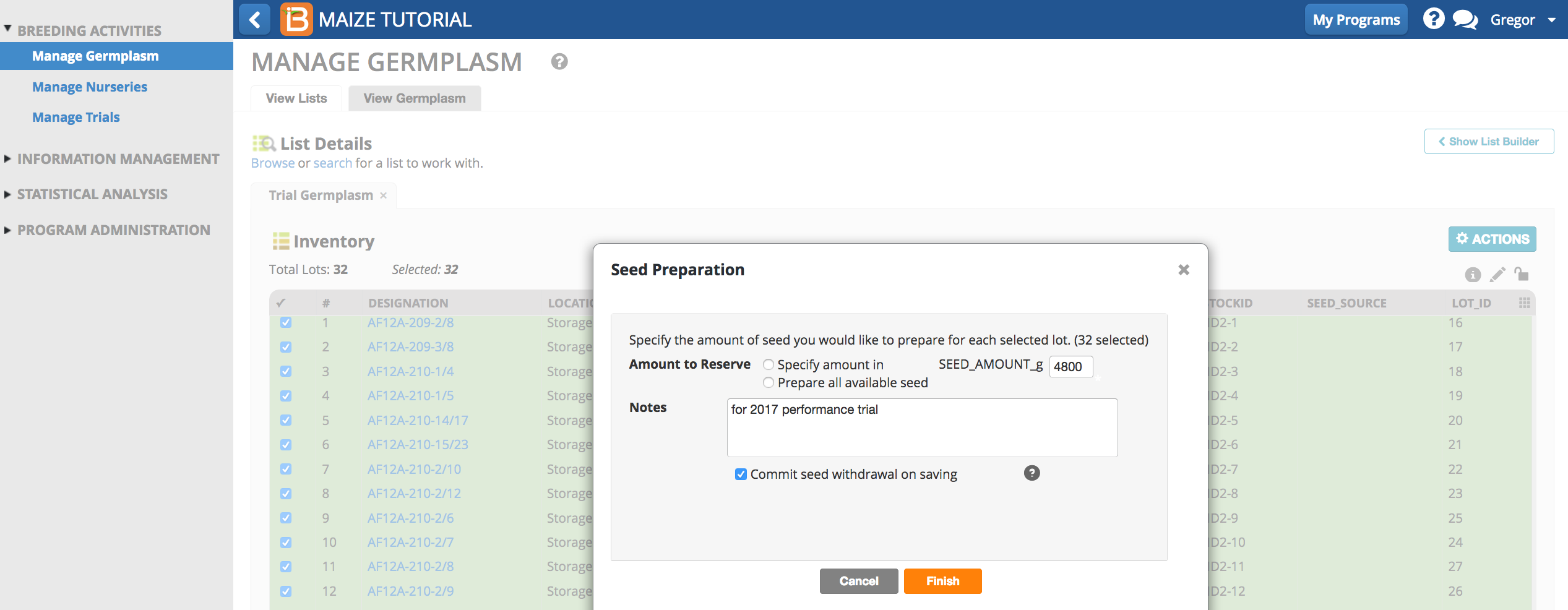
- Save changes.
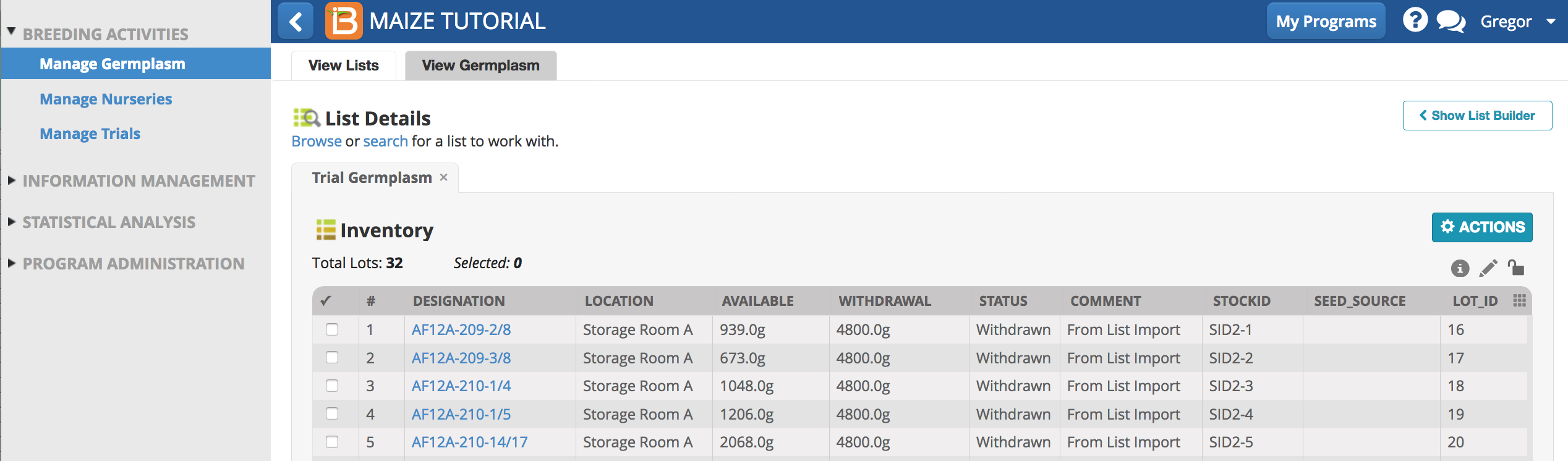
The inventory STATUS reads withdrawn for the amount of 4800g for each entry.
Related Materials
BMS Manual: Manage Trials
Maize Tutorial: Single Site Analysis: 4 Location Batch
Maize Tutorial: Multi-Site (GxE) Analysis
Maize Tutorial: Import Trial Data
The Integrated Breeding Platform (IBP) is jointly funded by: the Bill and Melinda Gates Foundation, the European Commission, United Kingdom's Department for International Development, CGIAR, the Swiss Agency for Development and Cooperation, and the CGIAR Fund Council. Coordinated by the Generation Challenge Program the Integrated Breeding Platform represents a diverse group of partners; including CGIAR Centers, national agricultural research institutes, and universities.
Maize ?demonstration data was provided by Mike Olsen at CIMMYT International Maize and Wheat Improvement Center. These data have been adapted for training purposes. Any misrepresentation of the raw breeding data is the solely the responsibility of the IBP.

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License?


